Call for Applications
Join us in shaping the future of musculoskeletal interface research
Our work bridges engineering, medicine, imaging, physics, biology, data science, computational modelling and machine learning to create new insights and technologies for musculoskeletal health. The RTG fosters a vibrant, collaborative, and international research environment – right in the heart of Hamburg’s rapidly growing life-science ecosystem.
The DFG Research Training Group “Multiscale Imaging and Analytics of Interfaces in Musculoskeletal Health” invites applications for fully funded doctoral positions starting in summer 2026.
We are looking for highly motivated candidates from biomedical engineering, medicine, physics, materials science, biology, data science, imaging, computer science, or related disciplines.
Your profile
We seek highly motivated and excellent candidates who:
- Hold an outstanding Master’s degree (or equivalent) in a discipline related to the specific project (e.g. biomedical engineering, physics, life sciences, data science, biomechanics, or related fields)
- Demonstrate a strong academic track record and a clear commitment to pursuing research at the highest international level
- Show proven interest and prior experience in at least one of the following areas: advanced imaging (X-ray, electron, light microscopy), experimental biomechanics, musculoskeletal biology, computational modeling, data science, or machine learning
- Show the capacity for independent, creative, and critical scientific thinking and are eager to take responsibility for a demanding doctoral research project
- Thrive in interdisciplinary research environments and value close collaboration across experimental, computational, and clinical domains
- Bring scientific curiosity, independence, initiative, and excellent communication skills
- Are committed to research integrity, open science, and fostering an inclusive and respectful research culture
We offer
- Fully funded 4-year doctoral positions within a structured and internationally visible DFG Research Training Group
- Intensive supervision by an interdisciplinary Supervisor Committee (SC) combining complementary expertise
- Access to state-of-the-art research infrastructure, including TEM, SBF-SEM, FIB-SEM, synchrotron beamlines, biomechanics laboratories, high-performance computing clusters, and advanced model systems
- A high-level qualification program tailored to ambitious doctoral researchers, featuring:
- Advanced imaging and data analytics modules
- A dedicated RTG lecture series by international experts
- Annual Winter Schools on Musculoskeletal Health and Interface Research
- Training in transferable skills, teaching, leadership, and career development
- Structured curricula in research data management and open science
- Strong support for scientific visibility, including conference participation, winter schools, and international research stays
- A stimulating, collaborative, and diversity-oriented research environment with family-friendly working conditions
- Targeted training in science communication and outreach, preparing you for careers in academia, industry, or beyond
Equality, Diversity & Inclusion (EDI)
We explicitly encourage women, first-generation academics, international applicants, and candidates from underrepresented groups to apply. Applicants with equal qualifications will be given preference according to DFG and institutional regulations for equity, diversity and inclusion (EDI). Applicants with families are strongly supported through UKE/TUHH family offices and flexible working arrangements.
Applications are accepted through the institutional recruiting websites, please click on the project of interest to be forwarded.
You can apply for a maximum of 3 projects!
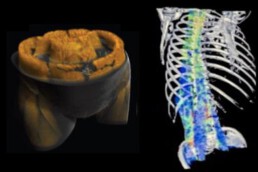
Supervisor: Isabel Molwitz (UKE)
Co-Supervisors: Tobias Knopp (TUHH/UKE) & Alexander Schlaefer (TUHH)
By focusing on spectral-CT imaging – an emerging technology increasingly available in radiology – this project will advance the predictive assessment of bone fragility. It explores the complex interactions between bone, muscle, and fatty degeneration of both, contributing to the development of more precise, non-invasive biomarkers for musculoskeletal health.
Specific aims
- Determine the predictive value of bone marrow fat quantified using spectral-CT techniques with enhanced AI-based image analysis for fracture risk, adverse outcome, and survival.
- Evaluate the complex interaction of bone, muscle, and fat contents for a more precise prediction of fracture risks, adverse outcomes, and reduced survival.
Profile of the applicant
- Master’s degree with excellent grades in computer science, related engineering fields, or applied mathematics
- Very good programming skills (Python, C++)
- Excellent knowledge of deep learning methods and frameworks, (deep) radiomics, and foundation models
- Expertise in medical imaging, medical image processing & deep learning, e.g., for image segmentation
- Excellent English and scientific writing skills
- Prior experience in academic publishing is highly regarded
P2 Exploring molecular pathways of tendon-to-bone healing in normal and osteoporotic bone remodeling
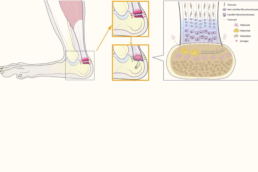
Supervisor: Johannes Keller (UKE)
Co-Supervisors: Katharina Jähn-Rickert (UKE) & Sara Checa (TUHH)
By providing new insights into the cellular and molecular mechanisms driving enthesis healing, bone remodeling, and tendon-bone attachment strength, this project addresses a critical challenge of regenerating MSK interfaces.
Specific aims
- Multiscale characterization of the impact of osteoporotic bone remodeling on tendon-to-bone and tendon-to-tendon healing (experimental).
- Explore the structure and cellular composition of the tendon-bone interface in patients with RC rerupture (clinical).
Profile of the applicant
- Strong interest and high motivation in addressing clinically relevant musculoskeletal research questions by translational approaches.
- Conscientious and structured work.
- Independence and flexibility, commitment, team and communication skills.
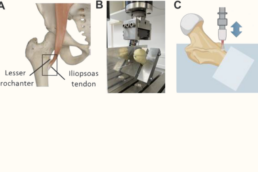
Supervisor: Benjamin Ondruschka (UKE)
Co-supervisors: Alexander Schlaefer & Sara Checa
This PhD project focuses on the biomechanical consequences of age-related degeneration at tendon–bone and muscle–tendon interfaces and their contribution to fracture risk and musculoskeletal fragility. The project integrates experimental biomechanics with tissue-level analyses to establish structure–function relationships in aging human musculoskeletal tissues.
Specific aims
- Quantify age-related structural and cellular changes in human tendon entheses, cortical bone, and myotendinous junctions
- Determine biomechanical properties and failure behavior of aging tendon–bone and muscle–tendon interfaces using uniaxial tensile testing and digital image correlation
- Correlate tissue microstructure with mechanical loading, force transmission, and fracture susceptibility
Profile of the applicant
- Master’s degree with excellent grades in Medical Sciences, Biomechanics, Biomedical or Mechanical Engineering, Movement Science, or a related field
- Strong background in experimental biomechanics or mechanical testing of biological tissues
- Basic knowledge of surgical anatomy, surgical techniques, autopsies, and post-mortem tissue handling
- High motivation for interdisciplinary research at the interface of biomechanics, medicine, and biology within an interdisciplinary research group project
- Excellent English and scientific writing skills; German is a plus
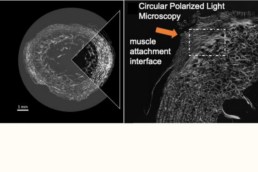
Supervisor: Björn Busse (UKE)
Co-Supervisors: Imke Greving (Hereon) & Benjamin Ondruschka (UKE)
By examining adaptations in enthesis and myotendinous junction (MTJ) in immobilized, physically active, and elderly individuals, this project will provide novel insights into the adaptive capacity of tendon interfaces. The findings will offer new multi-scale perspectives on interface integrity with implications for injury diagnoses, rehabilitation, and age-related musculoskeletal decline.
Specific aims
- Characterize the 2D/3D ultrastructure of mineralized and non-mineralized tissues at the enthesis and myotendinous junction to identify structural markers linked to interface integrity.
- Investigate microarchitectural differences across distinct physiological states, comparing immobilized individuals, physically active individuals, and elderly populations to understand how mechanical loading and aging influence enthesis adaptation and degeneration.
Profile of the applicant
- Outstanding Master’s degree, e.g. in biomedical engineering, life science, or other related fields
- Proven skills in high-resolution imaging and analytics (e.g., X-ray, confocal, electron microscopy, microCT, nanoCT, nanoindentation)
- Proven skills in 3D image and data analysis
- Experience in experimental and/or computational materials testing
- Previous experience in scientific presentation and publication
- High motivation for performing interdisciplinary research at the interface between experimental and computational biomechanics
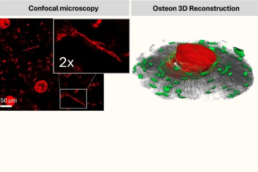
Supervisor: Katharina Jähn-Rickert (UKE)
Co-Supervisors: Maike Frye (UKE) & Tobias Knopp (UKE/ TUHH)
This project will provide novel insights into how osteocyte and vascular network impairments contribute to bone diseases such as osteoporosis, diabetic bone disease, and arteriosclerosis. A better understanding of structural homologies of these networks also with regard to pathological network disruptions can pave the way for targeted therapeutic approaches aimed at preserving network functions and osteocyte-vascular interactions, thereby improving bone health in pathological conditions.
Specific aims
- Assess pathological changes and identify structure-function alterations in osteocyte and vascular networks using dual-imaging approaches and AI-driven image processing.
- Determine structural mechanisms and processes involved in osteocyte and vascular network replenishment under pathological conditions.
Profile of the applicant
- Master degree in biological sciences, life sciences, biomedical engineering or related fields
- Interest in various biomedical imaging approaches
- Very good image processing skills
- Expertise in machine learning methods and data management advantageous
- Excellent English and scientific writing skills
- Prior experience with academic publishing or conference presentations is highly valued
Supervisor: Maike Frye (UKE)
Co-Supervisors: Imke Fiedler (UKE), Björn Busse (UKE)
The project will provide novel insights into how vascular dysfunction and endothelial mechanobiology influence bone-vessel interfaces. The findings will contribute to developing targeted therapeutic strategies to restore vascular integrity and improve bone health.
Specific aims
- Investigate the impact of bone matrix stiffness on endothelial cell behavior, focusing on mechanosensitive pathways in blood vessels.
- Determine how vascular dysfunction affects bone quality, exploring mechanisms that contribute to skeletal fragility and potential strategies for reversing these effects.
Profile of the applicant
- Master’s degree with excellent grades in molecular biology, biotechnology, biomedicine, biochemistry, or a related field
- Strong background in cardiovascular biology, extracellular matrix biology, or related areas
- Experience in animal experimentation (FELASA certification) is an advantage
- Highly motivated, able to work both independently and in a team, with strong organizational and coordination skills
- Excellent written and spoken English and strong scientific writing skills; German is a plus
- Prior experience with academic publishing is highly valued
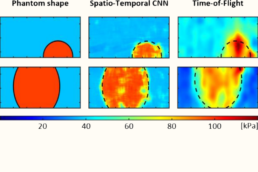
Supervisor: Alexander Schlaefer (TUHH)
Co-Supervisors: Checa (TUHH) & Ondruschka (UKE)
This project will establish deep learning-based methods to obtain quantitative estimates of soft tissue elasticity at high spatial and temporal resolution. The quantitative values could lead to markers of musculoskeletal health and form the basis for personalized biomechanical models.
Aim
- Establish suitable imaging parameters and machine learning architectures to estimate elastic tissue properties at interfaces in 2D, 3D and 4D from raw signals (Ultrasound/OCT) and with high resolution.
- Consider shear wave propagation at tissue interfaces and investigate state-of-the-art deep learning methods for detection and compensation of interface related artifacts.
Profile of the applicant
- Master’s degree with excellent grades in Computer Science, Applied Mathematics, or related fields.
- Very good programming skills (Python, C++)
- Excellent knowledge of machine learning methods and frameworks
- Expertise in robotics, medical imaging, medical image processing & machine learning; particularly for OCT and ultrasound
- Excellent English and scientific writing skills; German is a plus
- A collaborative mindset and prior experience in academic publishing
You will work on an interdisciplinary research project, to develop clinically applicable software to support physicians.
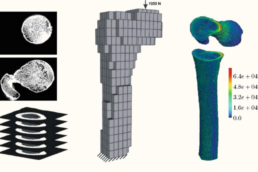
Supervisor: Alexander Düster (TUHH)
Co-Supervisors: Björn Busse (UKE) & Imke Fiedler (UKE)
By investigating the damping effect of marrow fat on vertebral bone mechanics, this study will provide deeper insights into skeletal biomechanics and fracture risk in metabolic bone diseases.
Aim
Develop a computational model based on CT scans and the Finite Cell Method to study the influence of fat on the mechanical damping behavior of vertebral bone.
Profile of the applicant
- Master’s degree with excellent grades in computational mechanics, computational engineering, mechanical engineering, biomedical engineering, or a closely related field.
- Demonstrated expertise in CT-based modeling and data-driven discretization for high-fidelity numerical simulations.
- Excellent knowledge of numerical simulation methods, ideally including the Finite Element Method, Finite Cell Method or comparable techniques.
- Prior experience with dynamic, time-domain simulations involving damping, multiphase, or viscoelastic behavior is advantageous.
- Excellent English and scientific writing skills; proficiency in German is a plus.
- Prior experience in academic publishing or clear potential for high-quality publications is highly valued.
- The successful candidate will possess excellent programming skills; a background in C++ for numerical simulation is highly desirable.
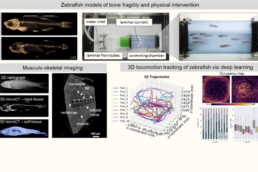
P9 Assessing musculoskeletal intervention using zebrafish locomotion analyses and tissue phenotyping
Supervisor: Imke Fiedler (UKE)
Co-Supervisors: Alexander Schlaefer (TUHH) & Maike Frye (UKE)
This project will establish a framework for correlating locomotion patterns with musculoskeletal disease phenotypes, enabling more efficient screening of disease mechanisms and interventions. This work will enhance our understanding of osteosarcopenic-vascular interactions and provide a scalable model for testing regenerative strategies.
Specific aims
- Establish zebrafish models with conditions impairing the MSK system for intelligent motion tracking and testing effects of exercise- and treatment-based intervention measures.
- Correlate MSK tissue phenotypes of zebrafish models with locomotion patterns, musculoskeletal activity, and swimming performance.
Profile of the applicant
- Master’s degree in biomedical engineering, life science, or other related fields
- Proven skills in high-resolution imaging and analytics (e.g., X-ray, confocal, electron microscopy, microCT, nanoCT, nanoindentation, synchrotron beamtime experience is advantageous)
- Proven skills in 3D image segmentation and quantification
- Skills in data analysis and statistics, basic Python knowledge is advantageous
- Experience with in vivo work with zebrafish models, ideally with a focus on musculoskeletal tissues, and 3R standards
- Highly motivated to work and collaborate at the interface between medicine, biology, imaging and data science
- Able to work both independently and in an interdisciplinary team, with strong organizational and coordination skills
- Excellent written and spoken English and strong scientific writing and presentation skills
- Interest in scientific outreach is highly valued
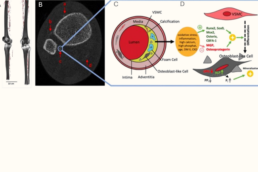
P10 What are the sex-specific vascular-driven mechanisms contributing to changes in gait and motion?
Supervisor: Felix von Brackel (UKE)
Co-Supervisors: Alexander Schlaefer (TUHH) & Maike Frye (UKE)
By integrating vascular imaging, neuromuscular testing, and computational modeling, this project will provide novel insights into how vascular alterations contribute to bone quality, movement impairments and fall risk.
Specific aims
- To characterize sex-specific gait and movement alterations associated with vascular calcification.
- To identify vascular and neuromuscular patterns that are associated with functional decline and increased fall risk.
- To investigate interactions between vascular health, neuromuscular performance, and bone quality using integrated imaging and functional data.
Profile of the applicant
- Master’s degree in engineering or informatics.
- Excellent knowledge of clinical imaging and image processing.
- Very good programming skills in Phyton.
- Expertise in medical imaging, image processing and anatomy.
- Databank skills including setup of databanks as well as merge of large datasets and its dedicated evaluation.
- Excellent skills in large cohort statistics.
- Expertise in data science.
- Knowledge of how to deal with patient data, anonymization and data handling.
- Excellent English and scientific writing skills; German skills is very welcome.
- Prior expertise in academic publishing is highly valued.
You will work in a highly interdisciplinary team including physicians and engineers. Your work is thought to enable physicians to tailor specific treatments and training plans for patients, based on the identified pattern. Furthermore, you will work in a clinical environment with contact to patients’ data and evaluations.